Circa
Gallery
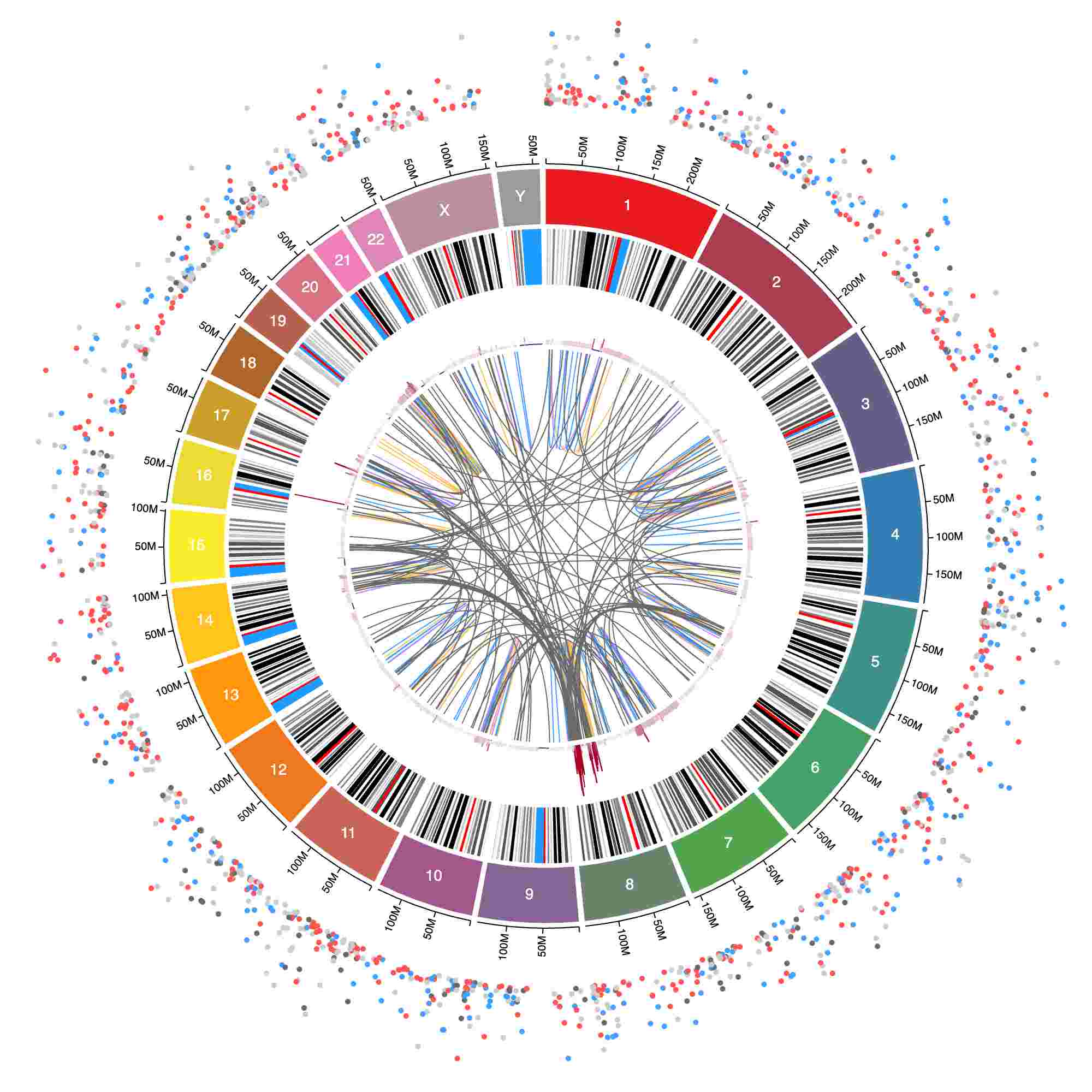
How to: Color by column
Coloring tracks by structural variant type
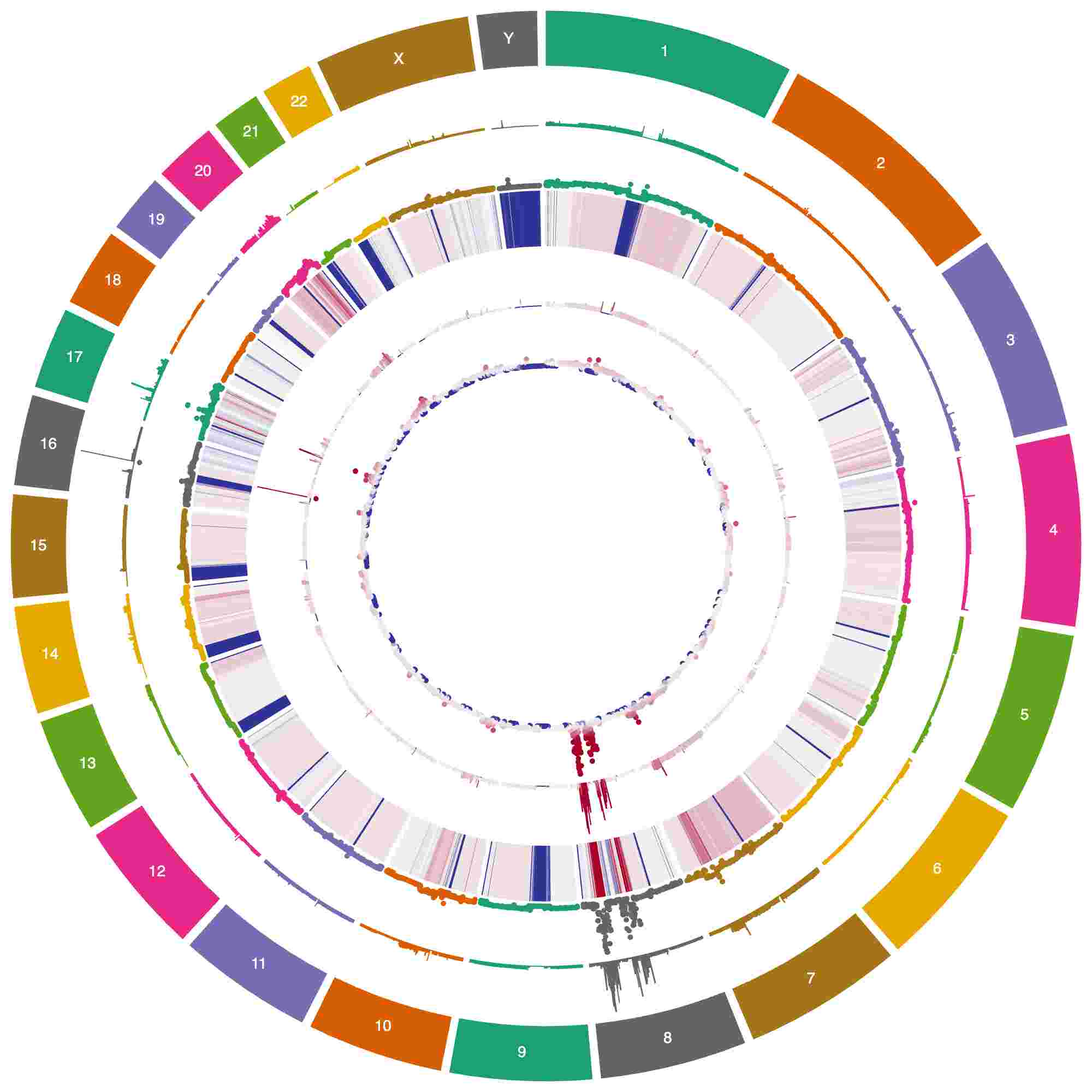
How to: Copy number ideas
Many ways to show copy number data in Circa
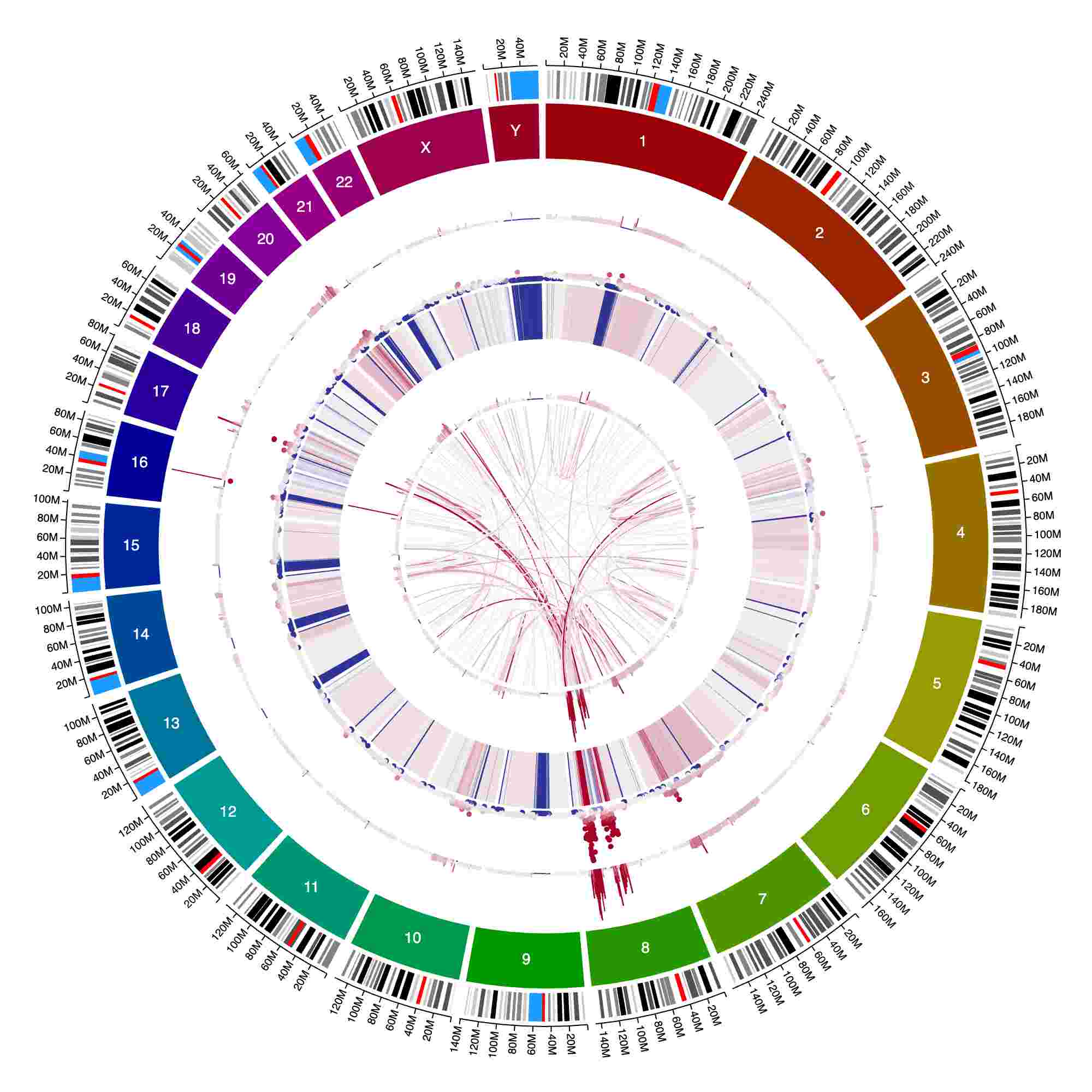
How to: Heatmap ideas
Several track types with continuous color scales
Documentation
- Supported input files
- Supported track types
- Data Mapper: Connecting data to a track
- Chromosome name matching
- Customizing chromosome names
- Flipping chromosomes and mirroring genomes
- Install Circa as an app
- Data privacy
- Register your license key