Flipping chromosomes and mirroring genomes
You can flip the orientation of chromosomes in Circa by adding a column to your reference setup dataset that indicates whether each chromosome should be flipped.
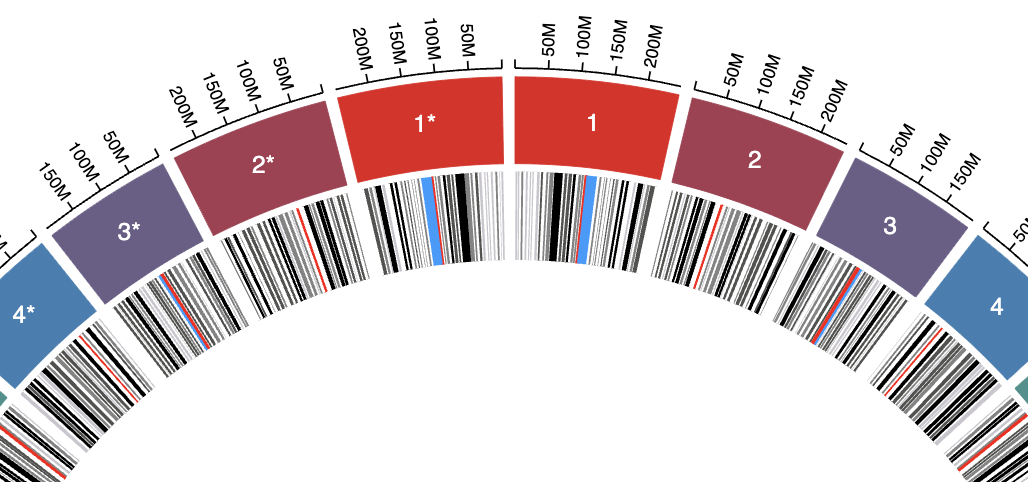
This is particularly useful when comparing two genomes to make them show up as if they are mirror images of each other.
Example setup
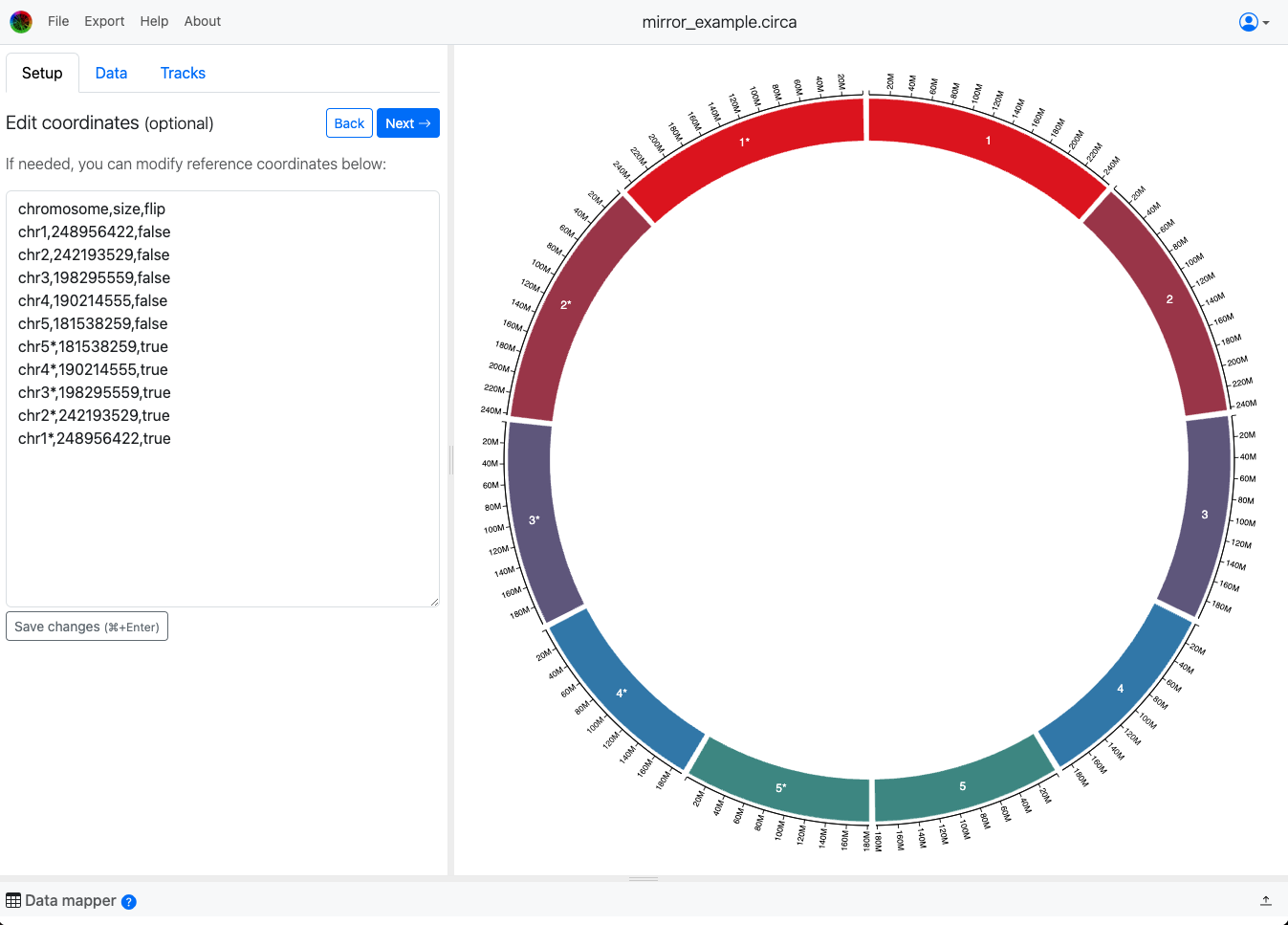
This is the CSV file used to make the example above:
chromosome,size,flip
chr1,248956422,false
chr2,242193529,false
chr3,198295559,false
chr4,190214555,false
chr5,181538259,false
chr5*,181538259,true
chr4*,190214555,true
chr3*,198295559,true
chr2*,242193529,true
chr1*,248956422,true
Note that chromosomes are still shown in the order they are listed, so if you want to mirror many chromosomes you would want to list the flipped ones in reverse order. See the example above where I did this for 5 chromosomes.
You can make this in Excel or Google Sheets and export as a CSV file.
The default "flip" status is `false`, so only the exact value "true" in that flip column will flip the chromosome. This works both when loading from a CSV/TSV file (recommended to save your work) or when writing it in the text area called "Edit coordinates" in the screenshot above.
How to set up
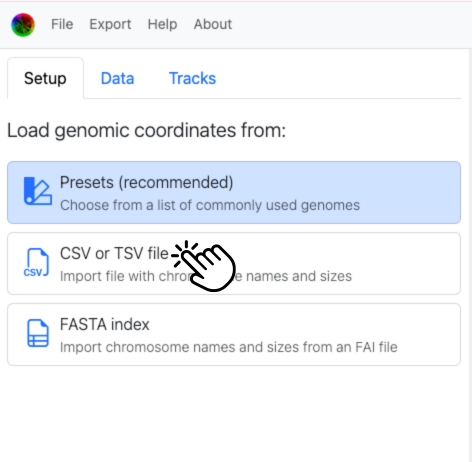
Choose CSV/TSV setup to include that flip column most easily:
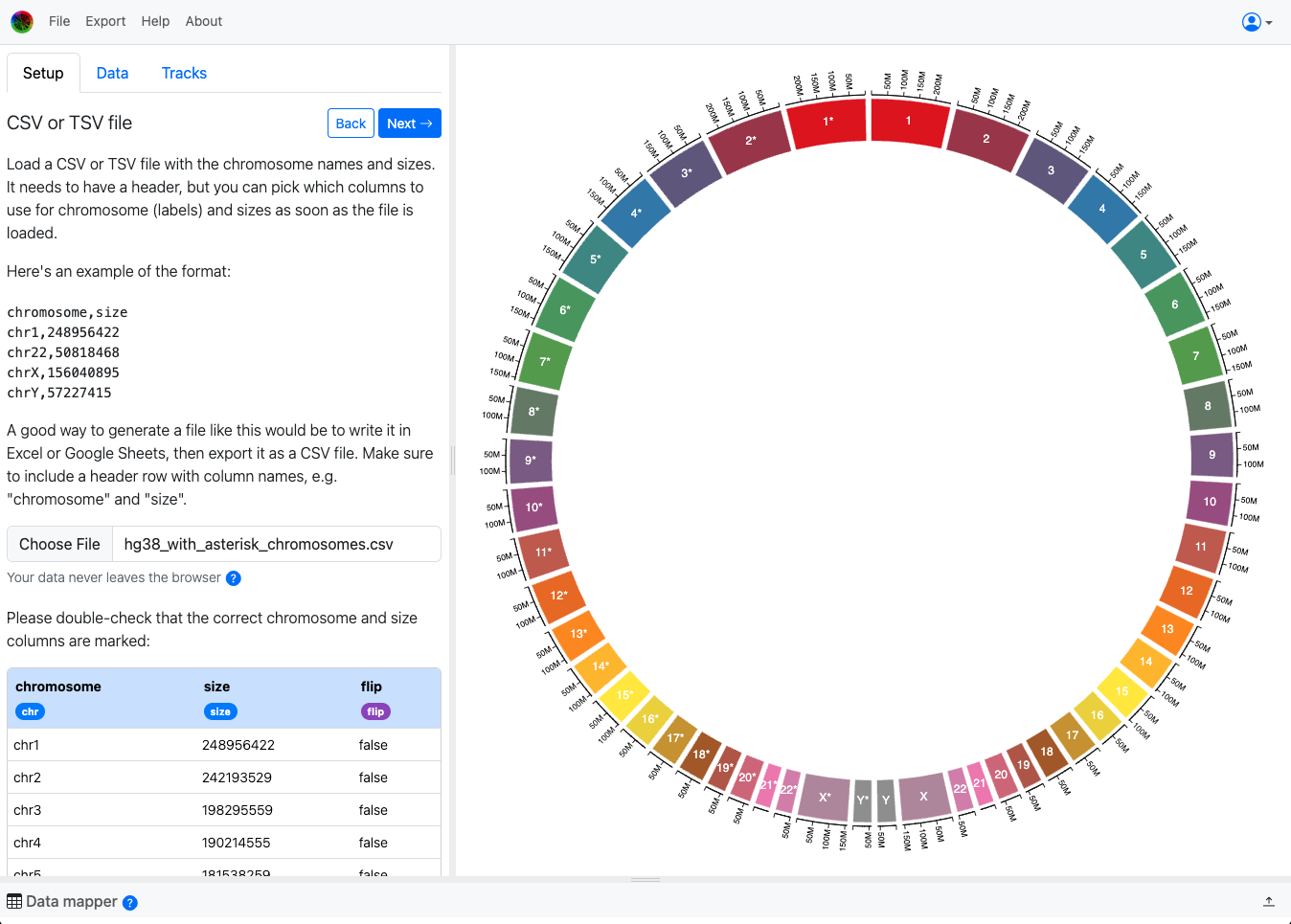
Then drag that purple flip tag onto the column that contains the flip information:
Plotting data onto these coordinates (after setup)
Important: The chromosome names MUST be unique. This is necessary because they are used as unique IDs for locating all the data points onto the right place in the coordinate system. So whatever names you use for the chromosomes, the data points need to use those same chromosome names.
Here is a small snippet of a file example where I copied the cytoband data and made a version that contains the names for the second set of chromosomes:
chromosome,start,end,locus,stain
chr1,0,2300000,p36.33,gneg
chr1,2300000,5300000,p36.32,gpos25
chr1*,0,2300000,p36.33,gneg
chr1*,2300000,5300000,p36.32,gpos25
Notice that all the data is the same, just duplicated to include the second set of chromosomes.
If you're not sure how to do that programmatically, we're happy to help. Just file a support ticket from the Help menu!
After modifying the whole cytoband TSV file like above and adding it in the Data tab, this is what we get:
Ta-da! If you have any questions, please don't hesitate to ask! Just click the Help menu and choose "Open support ticket".