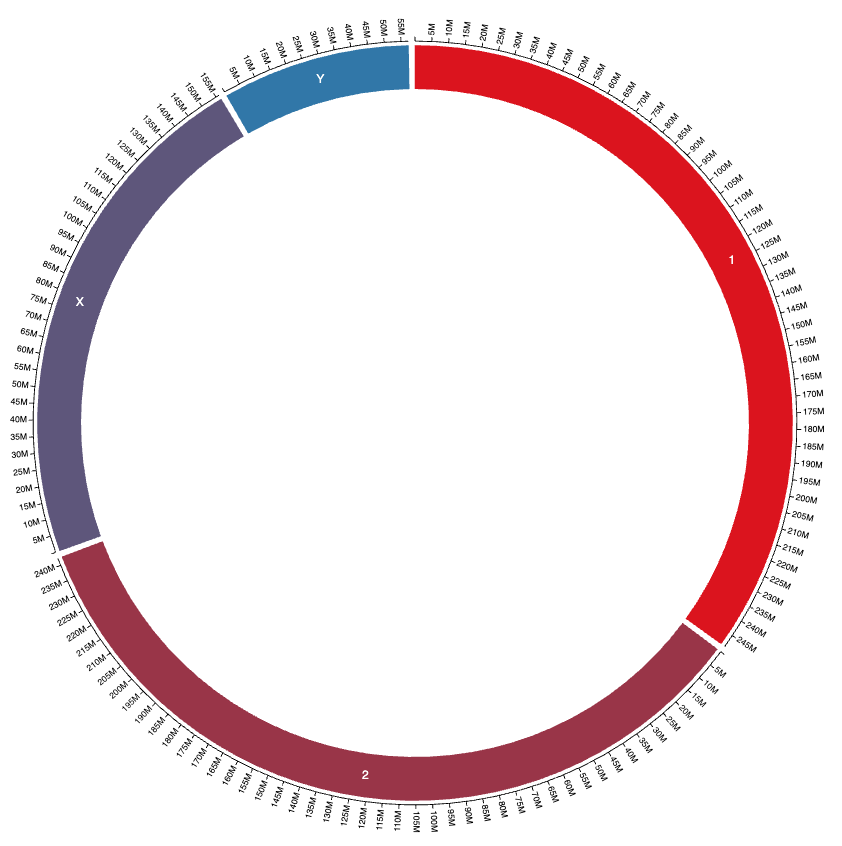
Customizing chromosome names
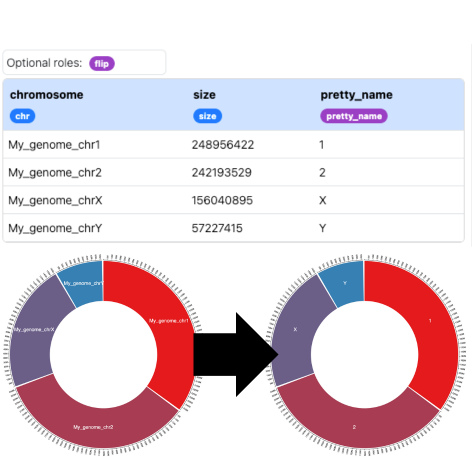
You can customize how chromosome names appear in the visualization by adding a "pretty_name" column to your reference setup dataset. This allows you to display more readable names while maintaining the original chromosome IDs for data mapping.
The pretty_name column is optional - if not provided, the chromosome name will be displayed as-is (with the chr prefix optionally hidden based on settings).
Example setup
Here is an example with ugly chromosome names:chromosome,size,pretty_name
My_genome_chr1,248956422,1
My_genome_chr2,242193529,2
My_genome_chrX,156040895,X
My_genome_chrY,57227415,Y
In the Setup tab, choose "CSV or TSV file" to load this file.
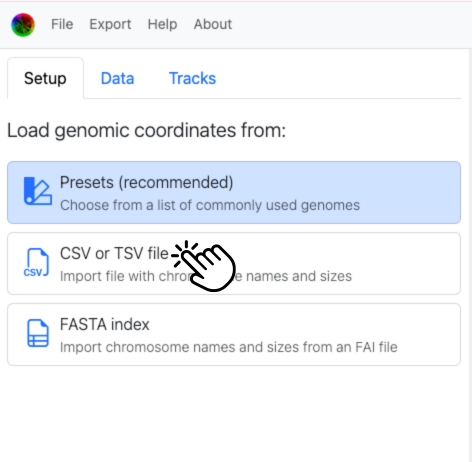
After loading this file, drag the purple pretty_name tag onto the column containing the display names (in this case it should already be there due to header-based guessing that we included to save you a step).
Important: The original chromosome names (not the pretty names) must still match between your reference setup and data files, since these are used as unique identifiers for placing data points.
And here is the result: